class: center, middle, inverse, title-slide .title[ # Data Visualisation and ggplot ] .author[ ### <br>Termeh Shafie ] --- layout: true --- class: middle ## What is in a dataset? --- ## Dataset terminology - Each row is an **observation** - Each column is a **variable** .small[ ```r starwars ``` ``` ## # A tibble: 87 × 14 ## name height mass hair_color skin_color eye_color birth_year ## <chr> <int> <dbl> <chr> <chr> <chr> <dbl> ## 1 Luke S… 172 77 blond fair blue 19 ## 2 C-3PO 167 75 <NA> gold yellow 112 ## 3 R2-D2 96 32 <NA> white, bl… red 33 ## 4 Darth … 202 136 none white yellow 41.9 ## 5 Leia O… 150 49 brown light brown 19 ## 6 Owen L… 178 120 brown, gr… light blue 52 ## # ℹ 81 more rows ## # ℹ 7 more variables: sex <chr>, gender <chr>, homeworld <chr>, ## # species <chr>, films <list>, vehicles <list>, ## # starships <list> ``` ] --- ## Luke Skywalker 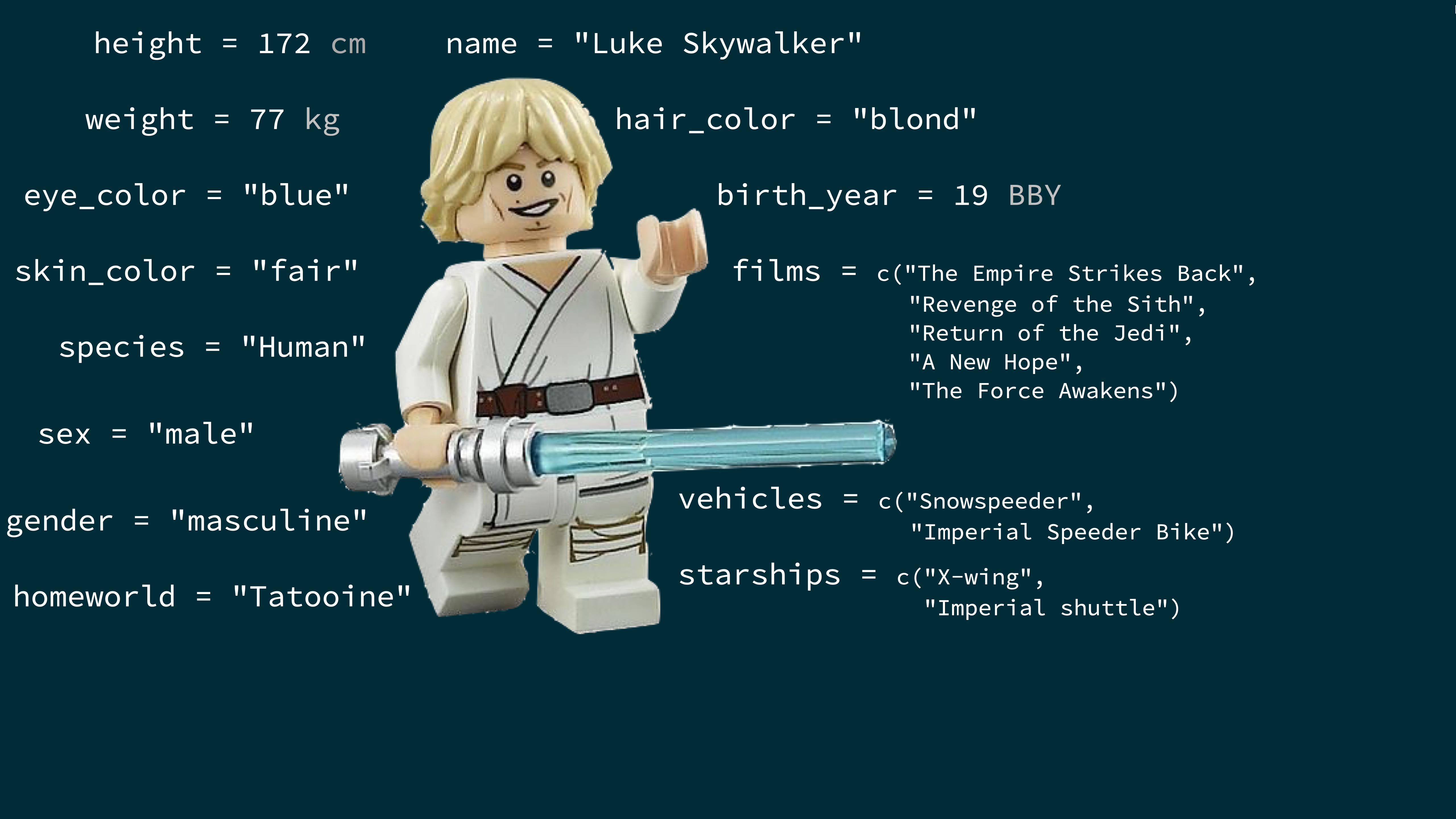 --- ## What's in the Star Wars data? Take a `glimpse` at the data: ```r glimpse(starwars) ``` ``` ## Rows: 87 ## Columns: 14 ## $ name <chr> "Luke Skywalker", "C-3PO", "R2-D2", "Darth V… ## $ height <int> 172, 167, 96, 202, 150, 178, 165, 97, 183, 1… ## $ mass <dbl> 77.0, 75.0, 32.0, 136.0, 49.0, 120.0, 75.0, … ## $ hair_color <chr> "blond", NA, NA, "none", "brown", "brown, gr… ## $ skin_color <chr> "fair", "gold", "white, blue", "white", "lig… ## $ eye_color <chr> "blue", "yellow", "red", "yellow", "brown", … ## $ birth_year <dbl> 19.0, 112.0, 33.0, 41.9, 19.0, 52.0, 47.0, N… ## $ sex <chr> "male", "none", "none", "male", "female", "m… ## $ gender <chr> "masculine", "masculine", "masculine", "masc… ## $ homeworld <chr> "Tatooine", "Tatooine", "Naboo", "Tatooine",… ## $ species <chr> "Human", "Droid", "Droid", "Human", "Human",… ## $ films <list> <"A New Hope", "The Empire Strikes Back", "… ## $ vehicles <list> <"Snowspeeder", "Imperial Speeder Bike">, <… ## $ starships <list> <"X-wing", "Imperial shuttle">, <>, <>, "TI… ``` --- .question[ How many rows and columns does this dataset have? What does each row represent? What does each column represent? ] ```r ?starwars ``` <img src="img/starwars-help.png" width="60%" style="display: block; margin: auto;" /> --- .question[ How many rows and columns does this dataset have? ] .pull-left[ ```r nrow(starwars) # number of rows ``` ``` ## [1] 87 ``` ```r ncol(starwars) # number of columns ``` ``` ## [1] 14 ``` ```r dim(starwars) # dimensions (row column) ``` ``` ## [1] 87 14 ``` ] --- class: middle ## Exploratory data analysis --- ## What is EDA? - Exploratory data analysis (EDA) is an approach to analysing data sets to summarize its main characteristics - Often, this is visual -- this is what we'll focus on first - But we might also calculate summary statistics and perform data wrangling/manipulation/transformation at (or before) this stage of the analysis -- this is what we'll focus on next --- ## Mass vs. height .question[ How would you describe the relationship between mass and height of Starwars characters? What other variables would help us understand data points that don't follow the overall trend? Who is the not so tall but really chubby character? ] <img src="data-viz_files/figure-html/unnamed-chunk-7-1.png" width="50%" style="display: block; margin: auto;" /> --- ## Jabba! <img src="img/jabbaplot.png" width="80%" style="display: block; margin: auto;" /> --- class: middle ## Data visualization --- ## Data visualization > *"The simple graph has brought more information to the data analyst's mind than any other device." --- John Tukey* - Data visualization is the creation and study of the visual representation of data - Many tools for visualizing data -- R is one of them - Many approaches/systems within R for making data visualizations -- **ggplot2** is one of them, and that's what we're going to use --- ## ggplot2 `\(\in\)` tidyverse .pull-left[ <img src="img/ggplot2-part-of-tidyverse.png" width="80%" style="display: block; margin: auto;" /> ] .pull-right[ - **ggplot2** is tidyverse's data visualization package - `gg` in "ggplot2" stands for Grammar of Graphics - Inspired by the book **Grammar of Graphics** by Leland Wilkinson ] --- ## Grammar of Graphics .pull-left-narrow[ A grammar of graphics is a tool that enables us to concisely describe the components of a graphic ] .pull-right-wide[ <img src="img/grammar-of-graphics.png" width="100%" style="display: block; margin: auto;" /> ] .footnote[ Source: [BloggoType](http://bloggotype.blogspot.com/2016/08/holiday-notes2-grammar-of-graphics.html)] --- ## Mass vs. height ```r ggplot(data = starwars, mapping = aes(x = height, y = mass)) + geom_point() + labs(title = "Mass vs. height of Starwars characters", x = "Height (cm)", y = "Weight (kg)") ``` ``` ## Warning: Removed 28 rows containing missing values or values outside the ## scale range (`geom_point()`). ``` <img src="data-viz_files/figure-html/mass-height-1.png" width="50%" style="display: block; margin: auto;" /> --- .question[ - What are the functions doing the plotting? - What is the dataset being plotted? - Which variables map to which features (aesthetics) of the plot? - What does the warning mean?<sup>+</sup> ] ```r ggplot(data = starwars, mapping = aes(x = height, y = mass)) + geom_point() + labs(title = "Mass vs. height of Starwars characters", x = "Height (cm)", y = "Weight (kg)") ``` ``` ## Warning: Removed 28 rows containing missing values or values outside the ## scale range (`geom_point()`). ``` .footnote[ <sup>+</sup>Suppressing warning to subsequent slides to save space ] --- ## Hello ggplot2! .pull-left-wide[ - `ggplot()` is the main function in ggplot2 - Plots are constructed in layers - Structure of the code for plots can be summarized as ````r ggplot(data = [dataset], mapping = aes(x = [x-variable], y = [y-variable])) + geom_xxx() + other options ```` - The ggplot2 package comes with the tidyverse ```r library(tidyverse) ``` - For help with ggplot2, see [ggplot2.tidyverse.org](http://ggplot2.tidyverse.org/) ] --- class: middle ## Why do we visualize? --- ## Anscombe's quartet .pull-left[ ``` ## set x y ## 1 I 10 8.04 ## 2 I 8 6.95 ## 3 I 13 7.58 ## 4 I 9 8.81 ## 5 I 11 8.33 ## 6 I 14 9.96 ## 7 I 6 7.24 ## 8 I 4 4.26 ## 9 I 12 10.84 ## 10 I 7 4.82 ## 11 I 5 5.68 ## 12 II 10 9.14 ## 13 II 8 8.14 ## 14 II 13 8.74 ## 15 II 9 8.77 ## 16 II 11 9.26 ## 17 II 14 8.10 ## 18 II 6 6.13 ## 19 II 4 3.10 ## 20 II 12 9.13 ## 21 II 7 7.26 ## 22 II 5 4.74 ``` ] .pull-right[ ``` ## set x y ## 23 III 10 7.46 ## 24 III 8 6.77 ## 25 III 13 12.74 ## 26 III 9 7.11 ## 27 III 11 7.81 ## 28 III 14 8.84 ## 29 III 6 6.08 ## 30 III 4 5.39 ## 31 III 12 8.15 ## 32 III 7 6.42 ## 33 III 5 5.73 ## 34 IV 8 6.58 ## 35 IV 8 5.76 ## 36 IV 8 7.71 ## 37 IV 8 8.84 ## 38 IV 8 8.47 ## 39 IV 8 7.04 ## 40 IV 8 5.25 ## 41 IV 19 12.50 ## 42 IV 8 5.56 ## 43 IV 8 7.91 ## 44 IV 8 6.89 ``` ] --- ## Summarising Anscombe's quartet ```r quartet %>% group_by(set) %>% summarise( mean_x = mean(x), mean_y = mean(y), sd_x = sd(x), sd_y = sd(y), r = cor(x, y) ) ``` ``` ## # A tibble: 4 × 6 ## set mean_x mean_y sd_x sd_y r ## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 I 9 7.50 3.32 2.03 0.816 ## 2 II 9 7.50 3.32 2.03 0.816 ## 3 III 9 7.5 3.32 2.03 0.816 ## 4 IV 9 7.50 3.32 2.03 0.817 ``` --- ## Visualizing Anscombe's quartet <img src="data-viz_files/figure-html/quartet-plot-1.png" width="80%" style="display: block; margin: auto;" /> --- class: middle # ggplot2 ❤️ 🐧 --- ## ggplot2 `\(\in\)` tidyverse .pull-left[ <img src="img/ggplot2-part-of-tidyverse.png" width="80%" style="display: block; margin: auto;" /> ] .pull-right[ - **ggplot2** is tidyverse's data visualization package - Structure of the code for plots can be summarized as ````r ggplot(data = [dataset], mapping = aes(x = [x-variable], y = [y-variable])) + geom_xxx() + other options ```` ] --- ## Data: Palmer Penguins Measurements for penguin species, island in Palmer Archipelago, size (flipper length, body mass, bill dimensions), and sex. .pull-left-narrow[ <img src="img/penguins.png" width="80%" style="display: block; margin: auto;" /> ] .pull-right-wide[ ```r library(palmerpenguins) glimpse(penguins) ``` ``` ## Rows: 344 ## Columns: 8 ## $ species <fct> Adelie, Adelie, Adelie, Adelie, Adeli… ## $ island <fct> Torgersen, Torgersen, Torgersen, Torg… ## $ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.… ## $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.… ## $ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195… ## $ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 362… ## $ sex <fct> male, female, female, NA, female, mal… ## $ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2… ``` ] --- .panelset[ .panel[.panel-name[Plot] <img src="data-viz_files/figure-html/unnamed-chunk-16-1.png" width="70%" style="display: block; margin: auto;" /> ] .panel[.panel-name[Code] ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + labs(title = "Bill depth and length", subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", x = "Bill depth (mm)", y = "Bill length (mm)", colour = "Species") ``` ``` ## Warning: Removed 2 rows containing missing values or values outside the ## scale range (`geom_point()`). ``` ] ] --- class: middle ## Coding out loud --- .midi[ > **Start with the `penguins` data frame** ] .pull-left[ ```r *ggplot(data = penguins) ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-17-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > **map bill depth to the x-axis** ] .pull-left[ ```r ggplot(data = penguins, * mapping = aes(x = bill_depth_mm)) ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-18-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > map bill depth to the x-axis > **and map bill length to the y-axis.** ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, * y = bill_length_mm)) ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-19-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > map bill depth to the x-axis > and map bill length to the y-axis. > **Represent each observation with a point** ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm)) + * geom_point() ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-20-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > map bill depth to the x-axis > and map bill length to the y-axis. > Represent each observation with a point > **and map species to the colour of each point.** ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, * colour = species)) + geom_point() ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-21-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > map bill depth to the x-axis > and map bill length to the y-axis. > Represent each observation with a point > and map species to the colour of each point. > **Title the plot "Bill depth and length"** ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + * labs(title = "Bill depth and length") ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-22-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > map bill depth to the x-axis > and map bill length to the y-axis. > Represent each observation with a point > and map species to the colour of each point. > Title the plot "Bill depth and length", > **add the subtitle "Dimensions for Adelie, Chinstrap, and Gentoo Penguins"** ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + labs(title = "Bill depth and length", * subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins") ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-23-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > map bill depth to the x-axis > and map bill length to the y-axis. > Represent each observation with a point > and map species to the colour of each point. > Title the plot "Bill depth and length", > add the subtitle "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", > **label the x and y axes as "Bill depth (mm)" and "Bill length (mm)", respectively** ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + labs(title = "Bill depth and length", subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", * x = "Bill depth (mm)", y = "Bill length (mm)") ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-24-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > map bill depth to the x-axis > and map bill length to the y-axis. > Represent each observation with a point > and map species to the colour of each point. > Title the plot "Bill depth and length", > add the subtitle "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", > label the x and y axes as "Bill depth (mm)" and "Bill length (mm)", respectively, > **label the legend "Species"** ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + labs(title = "Bill depth and length", subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", x = "Bill depth (mm)", y = "Bill length (mm)", * colour = "Species") ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-25-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > map bill depth to the x-axis > and map bill length to the y-axis. > Represent each observation with a point > and map species to the colour of each point. > Title the plot "Bill depth and length", > add the subtitle "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", > label the x and y axes as "Bill depth (mm)" and "Bill length (mm)", respectively, > label the legend "Species", > **and add a caption for the data source.** ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + labs(title = "Bill depth and length", subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", x = "Bill depth (mm)", y = "Bill length (mm)", colour = "Species", * caption = "Source: Palmer Station LTER / palmerpenguins package") ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-26-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .midi[ > Start with the `penguins` data frame, > map bill depth to the x-axis > and map bill length to the y-axis. > Represent each observation with a point > and map species to the colour of each point. > Title the plot "Bill depth and length", > add the subtitle "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", > label the x and y axes as "Bill depth (mm)" and "Bill length (mm)", respectively, > label the legend "Species", > and add a caption for the data source. > **Finally, use a discrete colour scale that is designed to be perceived by viewers with common forms of colour blindness.** ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + labs(title = "Bill depth and length", subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", x = "Bill depth (mm)", y = "Bill length (mm)", colour = "Species", caption = "Source: Palmer Station LTER / palmerpenguins package") + * scale_colour_viridis_d() ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-27-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .panelset[ .panel[.panel-name[Plot] <img src="data-viz_files/figure-html/unnamed-chunk-28-1.png" width="70%" style="display: block; margin: auto;" /> ] .panel[.panel-name[Code] ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + labs(title = "Bill depth and length", subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", x = "Bill depth (mm)", y = "Bill length (mm)", colour = "Species", caption = "Source: Palmer Station LTER / palmerpenguins package") + scale_colour_viridis_d() ``` ``` ## Warning: Removed 2 rows containing missing values or values outside the ## scale range (`geom_point()`). ``` ] .panel[.panel-name[Narrative] .pull-left-wide[ .midi[ Start with the `penguins` data frame, map bill depth to the x-axis and map bill length to the y-axis. Represent each observation with a point and map species to the colour of each point. Title the plot "Bill depth and length", add the subtitle "Dimensions for Adelie, Chinstrap, and Gentoo Penguins", label the x and y axes as "Bill depth (mm)" and "Bill length (mm)", respectively, label the legend "Species", and add a caption for the data source. Finally, use a discrete colour scale that is designed to be perceived by viewers with common forms of colour blindness. ] ] ] ] --- ## Argument names .tip[ You can omit the names of first two arguments when building plots with `ggplot()`. ] .pull-left[ ```r ggplot(data = penguins, mapping = aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + scale_colour_viridis_d() ``` ] .pull-right[ ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, colour = species)) + geom_point() + scale_colour_viridis_d() ``` ] --- class: middle ## Aesthetics --- ## Aesthetics options Commonly used characteristics of plotting characters that can be **mapped to a specific variable** in the data are - `colour` - `shape` - `size` - `alpha` (transparency) --- ## Colour .pull-left[ ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, * colour = species)) + geom_point() + scale_colour_viridis_d() ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-29-1.png" width="100%" style="display: block; margin: auto;" /> ] --- ## Shape Mapped to a different variable than `colour` .pull-left[ ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, colour = species, * shape = island)) + geom_point() + scale_colour_viridis_d() ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-30-1.png" width="100%" style="display: block; margin: auto;" /> ] --- ## Shape Mapped to same variable as `colour` .pull-left[ ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, colour = species, * shape = species)) + geom_point() + scale_colour_viridis_d() ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-31-1.png" width="100%" style="display: block; margin: auto;" /> ] --- ## Size .pull-left[ ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, colour = species, shape = species, * size = body_mass_g)) + geom_point() + scale_colour_viridis_d() ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-32-1.png" width="100%" style="display: block; margin: auto;" /> ] --- ## Alpha .pull-left[ ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, colour = species, shape = species, size = body_mass_g, * alpha = flipper_length_mm)) + geom_point() + scale_colour_viridis_d() ``` ] .pull-right[ <img src="data-viz_files/figure-html/unnamed-chunk-33-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .pull-left[ **Mapping** ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm, * size = body_mass_g, * alpha = flipper_length_mm)) + geom_point() ``` <img src="data-viz_files/figure-html/unnamed-chunk-34-1.png" width="100%" style="display: block; margin: auto;" /> ] .pull-right[ **Setting** ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm)) + * geom_point(size = 2, alpha = 0.5) ``` <img src="data-viz_files/figure-html/unnamed-chunk-35-1.png" width="100%" style="display: block; margin: auto;" /> ] --- ## Mapping vs. setting - **Mapping:** Determine the size, alpha, etc. of points based on the values of a variable in the data - goes into `aes()` - **Setting:** Determine the size, alpha, etc. of points **not** based on the values of a variable in the data - goes into `geom_*()` (this was `geom_point()` in the previous example, but we'll learn about other geoms soon!) --- class: middle ## Faceting --- ## Faceting - Smaller plots that display different subsets of the data - Useful for exploring conditional relationships and large data --- .panelset[ .panel[.panel-name[Plot] <img src="data-viz_files/figure-html/unnamed-chunk-36-1.png" width="70%" style="display: block; margin: auto;" /> ] .panel[.panel-name[Code] ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm)) + geom_point() + * facet_grid(species ~ island) ``` ``` ## Warning: Removed 2 rows containing missing values or values outside the ## scale range (`geom_point()`). ``` ] ] --- ## Various ways to facet .question[ In the next few slides describe what each plot displays. Think about how the code relates to the output. **Note:** The plots in the next few slides do not have proper titles, axis labels, etc. because we want you to figure out what's happening in the plots. But you should always label your plots! ] --- ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm)) + geom_point() + * facet_grid(species ~ sex) ``` <img src="data-viz_files/figure-html/unnamed-chunk-37-1.png" width="60%" style="display: block; margin: auto;" /> --- ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm)) + geom_point() + * facet_grid(sex ~ species) ``` <img src="data-viz_files/figure-html/unnamed-chunk-38-1.png" width="60%" style="display: block; margin: auto;" /> --- ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm)) + geom_point() + * facet_wrap(~ species) ``` <img src="data-viz_files/figure-html/unnamed-chunk-39-1.png" width="60%" style="display: block; margin: auto;" /> --- ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm)) + geom_point() + * facet_grid(. ~ species) ``` <img src="data-viz_files/figure-html/unnamed-chunk-40-1.png" width="60%" style="display: block; margin: auto;" /> --- ```r ggplot(penguins, aes(x = bill_depth_mm, y = bill_length_mm)) + geom_point() + * facet_wrap(~ species, ncol = 2) ``` <img src="data-viz_files/figure-html/unnamed-chunk-41-1.png" width="60%" style="display: block; margin: auto;" /> --- ## Faceting summary - `facet_grid()`: - 2d grid - `rows ~ cols` - use `.` for no split - `facet_wrap()`: 1d ribbon wrapped according to number of rows and columns specified or available plotting area --- ## Facet and color .pull-left-narrow[ ```r ggplot( penguins, aes(x = bill_depth_mm, y = bill_length_mm, * color = species)) + geom_point() + facet_grid(species ~ sex) + * scale_color_viridis_d() ``` ] .pull-right-wide[ <img src="data-viz_files/figure-html/unnamed-chunk-42-1.png" width="100%" style="display: block; margin: auto;" /> ] --- ## Face and color, no legend .pull-left-narrow[ ```r ggplot( penguins, aes(x = bill_depth_mm, y = bill_length_mm, color = species)) + geom_point() + facet_grid(species ~ sex) + scale_color_viridis_d() + * guides(color = "none") ``` ] .pull-right-wide[ <img src="data-viz_files/figure-html/unnamed-chunk-43-1.png" width="100%" style="display: block; margin: auto;" /> ] --- .your-turn[ .light-blue[.hand[Practical:]] `Starwars + Quarto` - Visualizing Starwars characters <img src="img/luke-skywalker.png" width="75%" style="display: block; margin: auto;" /> ]